Center for Computational Biology
Department of Molecular Biosciences
University of Kansas
Research: Integrative Theories and Experiments in Living Systems
In the Ray lab we are working to discover new ways of understanding how molecular networks affect biological growth, fitness, and evolution. We use a theory-driven experimental approach, with a foundation in systems biology, chemical and statistical physics, and information theory. Experimental synthetic biology and single-cell measurements provide an empirical foundation for our theoretical and computational work.
One of the major goals of our lab is to provide a generalized theoretical framework for understanding how infectious diseases and cancer cell growth are guided by molecular mechanisms, how they evolve, and how their growth predisposes them to novel treatment strategies. To this end we are developing experimental and theoretical approaches to predict when cells enter antibiotic-tolerant states, characterizing the statistics of how such states are inherited, and determining the selective pressures on cells on the threshold of critical transitions. The figure below shows an example of a recent effort in the lab toward these goals.
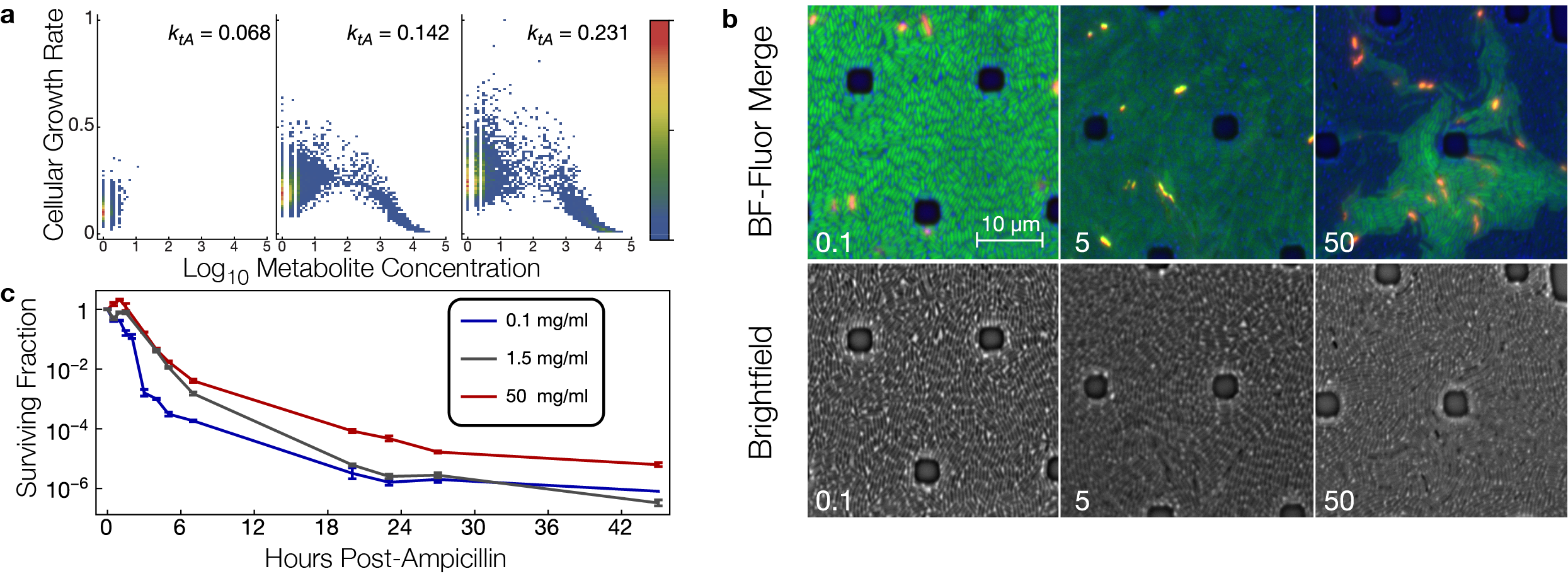
Simulations and experiments on a metabolic step affecting cellular growth rates in bacteria. a. Stochastic simulations of a metabolic step. Growth feedback from metabolite toxicity traps a subset of trajectories into irreversible metabolite buildup with consequent cellular growth arrest. The parameter ktA affects the rate at which precursor metabolite enters the simulated system. b. Microscopy of Escherichia coli cells (strain B REL606 lacI– PlacO1GFP) grown in a microfluidic device with a constant flow of minimal medium containing lactose at the indicated concentration (in mg/ml). Red and yellow cells are stained with propidium iodide, which indicates cell death. When growth-mediated dilution of green fluorescent protein (GFP) is slowed or halted, the cells are brighter. Brighter green cells are slower growing. At high lactose concentrations, cellular growth rates are heterogeneous, consistent with the simulations predicting growth arrest in a subset of cells. c. Emergence of growth arrested cells in high lactose concentrations is correlated with enrichment of antibiotic persister cells in these conditions, as indicated by the higher fraction of long-lived cells surviving in ampicillin.
Research Profiles of Scientists in the Lab
Shamus Cooley: Analysis of High-Dimensional Datasets

Cells carry a lot of information about their identity, such as their cell type or phenotype. We are using high-dimensional data, especially single-cell RNA-Sequencing, to computationally characterize cellular identity and differentiation dynamics with a machine learning approach.
GW McElfresh: Bacterial Population Heterogeneity

My work involves using computational and experimental tools to study gene regulatory and cellular developmental programs involved in cellular heterogeneity and inter-generational information transfer. To study bacterial growth transitions, I am using tools from image analysis to more robustly observe Escherichia coli cells as they grow, diversify, and undergo phenotypic changes such as becoming a persister cell. I am also using RNA sequencing data to investigate global gene expression shifts and identifying root causes of phenotypic changes.
Adam Podgorny: Predicting Cell Cycle Dynamics

Control of the cell cycle is like conducting an orchestra where each instrument is partially random, and getting it to play coherent music anyway. My project is to analyze and predict the life cycle of a cell in space and time. My first goal is to predict how chromosome segregation interacts with cell growth in bacteria. To that end, I am developing a coarse-grained spatial chromosome simulator designed to interact with our lab's cell cycle framework.
Huijing Wang: Information Transfer and Prediction of Phenotypic Transitions

Pathogenic bacteria can cause acute or chronic infections in humans. Acute infection is generally treatable with antibiotic therapy, but immune responses and antibiotics can push bacterial cells into a slow-growing state with increased antibiotic persistence. Many slow-growing infectious bacteria can form biofilms and cause chronic or stubborn diseases that are recalcitrant to treatment, such as Pseudomonas aeruginosa infection in cystic fibrosis patients and urinary tract infections by uropathogenic Escherichia coli.
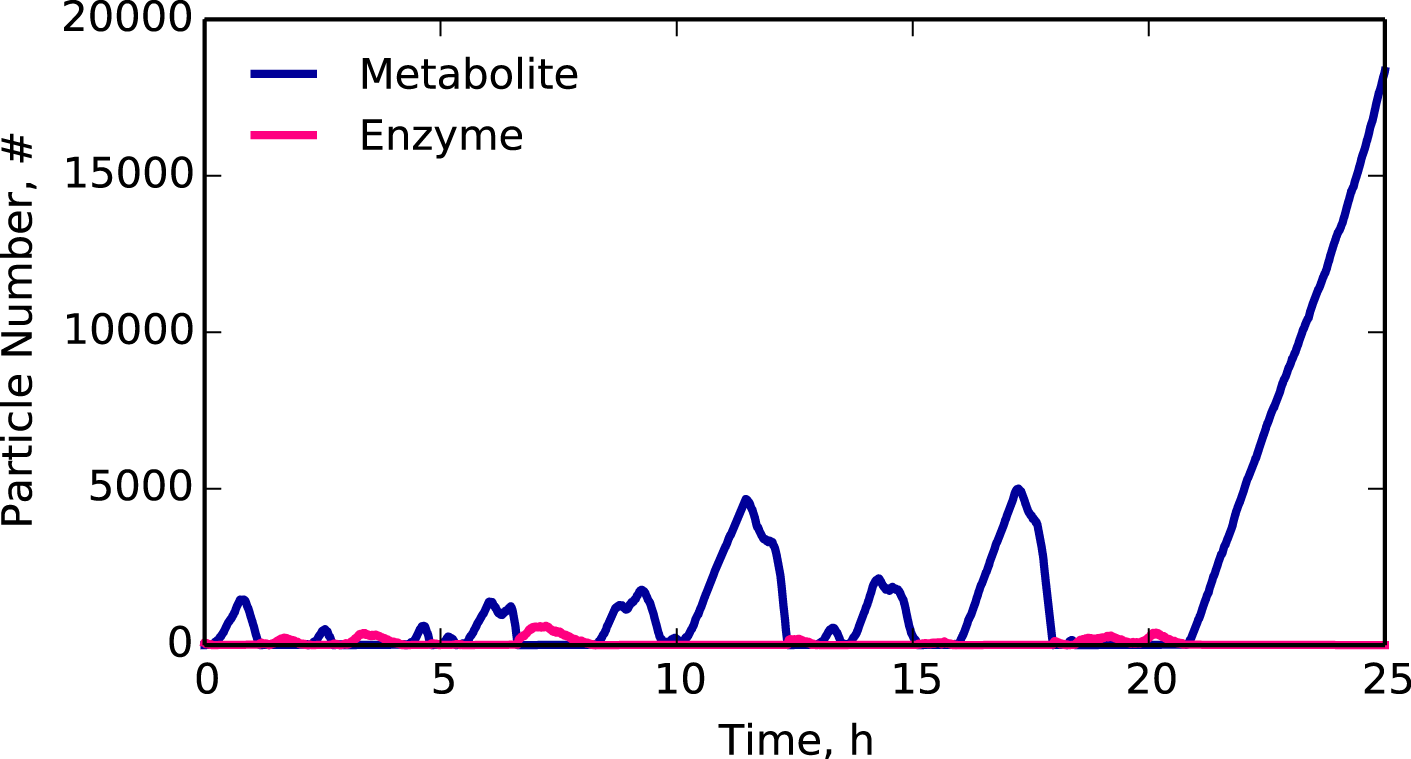
Simulated trajectory of a metabolite on the edge of a critical transition.
Can we predict when cells transition to growth arrest?
In ecological and economic studies, imminent transitions often display dynamical signatures that act as early warning signs. Based on this observation, we are testing the hypothesis that molecular networks in cells exhibit similar early warning signs.
Nishantha Wijesuriya: Dynamics of Single-Cell Gene Expression

We are developing methods to study gene expression programs in response to stress.